Structural Biology- Our interests are to determine the structure/function relationships driving biological process. We are particularly interested in the molecular recognition events that lead to ligand- induced receptor activation and regulation. We also have programs in structure determination of membrane proteins and multi-protein complexes.
Protein Engineering and Biophysical Characterization- We use protein engineering to introduce and modify changes in protein structures in order to develop new protein functions or to systematically evaluate the roles of specific protein groups on the overall function. Biophysical characterization is used to provide a picture of the energy landscape governing affinity and specificity in protein-protein interactions.
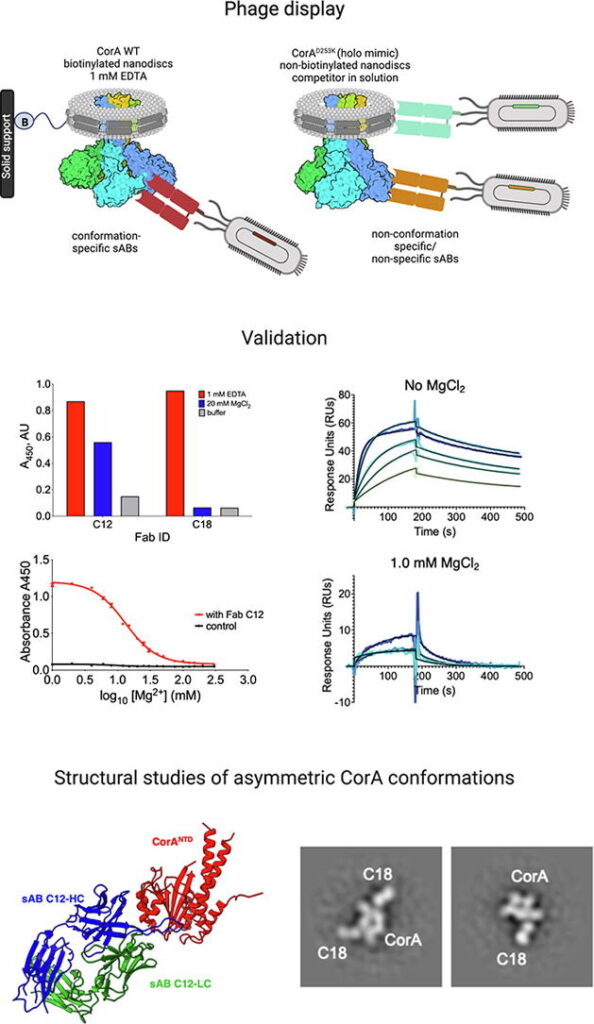
Synthetic Antibodies- We use powerful phage display libraries to generate novel antibody-like molecules called “Synthetic Antigen Binders” (sABs) that can be “evolved” to bind tightly and specifically to a broad range of targets including soluble proteins, multi-protein complexes, membrane proteins and functional RNAs. sABs have multiple purposes as described below.
Hormone-induced receptor activation and regulation- is driven by an intricate set of protein-protein interactions. We study the role of these sophisticated molecular recognition events on the activities of a series of cytokine hormone-receptor complexes. Using the human pituitary hormones: growth hormone (hGH), prolactin (hPRL) and placental lactogen (hPL) as key model systems, we have developed one of the most comprehensive analyses of the structure-function relationships governing protein-protein interactions in general.
Comprehensive and quantitative mapping of energy landscapes- for protein-protein interactions by a rapid combinatorial scanning. We have developed a high throughput quantitative saturation (QS) scanning strategy to obtain a comprehensive data base of all possible mutations across a large protein-protein interface. Using this method, we were able to substitute all 20 amino acid types at 35 positions (700 mutations) in hGH and evaluate the relative changes in ddG of all the substitutions. This work represents the most extensive evaluation of a protein-protein interface has ever been undertaken and provides a complete view of the energy landscape resulting in altering some important elements of the current molecular recognition dogma.
Principal determinates leading to the transition-state structure- for formation of protein-protein complexes. The binding transition state (TS) is the rate determining step in all molecular recognition processes, but was only understood at a very qualitative level. To establish a more quantitative picture of the TS, we exploited a set of biophysical techniques that have been widely used in protein folding studies. These evaluations led to a picture of the TS structure where the two interacting surfaces attain an orientational complementary by the time the TS is reached.
Chaperone-Assisted Crystallography (CAC)- A majority of the most high-value structure biology targets remain unsolved even after heroic efforts. To overcome the barriers to crystallization, we have developed a strategy using crystallization chaperones. These chaperones take the form of antigen-binding fragments (Fabs) that are engineered by phage display mutagenesis to bind to the target proteins. We have shown that chaperones can promote crystallization by both stabilizing the target proteins and providing primary contacts between molecules in the crystalline lattice. A high throughput pipeline has been assembled that will provide multiple unique chaperones for each designated target. We have shown that this approach can be used to successfully crystallize difficult targets like multiprotein complexes, membrane proteins and RNA.
Receptor-mediated drug delivery (RMD)- Directed delivery of bioactive reagents into cells is one of the most intensely pursued objectives in biomedical research, yet technical barriers have restricted progress. The fundamental goal is to develop a technology to transport reagents across the cell membrane that can have a direct influence on cellular functions in diseased cells, while sparing normal ones. To this end, we have developed an efficient and robust methodology that facilitates the delivery of protein cargos into the cytoplasm of cancer cells, leaving normal cells unaffected. Our approach exploits the ligand-induced receptor-mediated internalization pathways as extremely efficient and selective transmembrane delivery vehicles. Our approach employs the judicious selection of the ligand-receptor systems that are extremely versatile in their ability to deliver cargos of different size and chemical composition.
