Search for author, keywords, or any other term.
Erramilli, Satchal K; Nosol, Kamil; Pietrzak-Lichwa, Krzysztof; Schmandt, Nicolaus; Li, Tian; Tokarz, Piotr; Hou, Jingkai; Zhao, Minglei; Perozo, Eduardo; Kossiakoff, Anthony A
Conformational ensembles of the magnesium channel CorA reveal structural basis for channel gating Journal Article
In: Proc Natl Acad Sci U S A, vol. 123, no. 8, pp. e2512532123, 2026, ISSN: 1091-6490.
@article{pmid41701836,
title = {Conformational ensembles of the magnesium channel CorA reveal structural basis for channel gating},
author = {Satchal K Erramilli and Kamil Nosol and Krzysztof Pietrzak-Lichwa and Nicolaus Schmandt and Tian Li and Piotr Tokarz and Jingkai Hou and Minglei Zhao and Eduardo Perozo and Anthony A Kossiakoff},
doi = {10.1073/pnas.2512532123},
issn = {1091-6490},
year = {2026},
date = {2026-02-01},
urldate = {2026-02-01},
journal = {Proc Natl Acad Sci U S A},
volume = {123},
number = {8},
pages = {e2512532123},
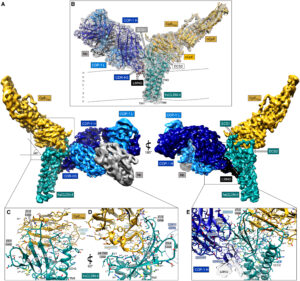
abstract = {In prokaryotes, CorA is the primary influx pathway for magnesium, a critical divalent cation in cellular physiology and biochemistry. Mechanistic studies show that homopentameric CorA is regulated through an intracellular [Mg]-dependent negative feedback loop, involving the asymmetric participation of individual subunits. To understand the connection between asymmetry and activation, we used single-particle cryo-EM to solve sixteen structures of nanodisc-reconstituted CorA. We utilized conformation-specific synthetic antibodies to stabilize subtle but significant conformational differences in the cryo-EM structures. Our results demonstrate that CorA exists as a set of conformational ensembles, where population size inversely correlates with intracellular Mg concentration. These ensembles include channels with a variety of pore conformations, both constricted and dilated, suggesting a spectrum of active CorA functional states. The ensembles connect asymmetric structural transitions in the cytoplasmic domain with conformational changes in the permeation pathway via an electrostatic network, ultimately controlling channel-gating events. We believe that these results establish a framework for understanding magnesium homeostasis in prokaryotic systems.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

Han, Chengxiao; Weng, Yicheng; Zheng, Qingyun; Qu, Qian; Erramilli, Satchal K; Su, Zhen; Duan, Yujuan; Han, Yunxi; Zhai, Xiaoguo; Li, Jingxian; Kossiakoff, Anthony A; Pan, Man; Zhao, Minglei; Liu, Lei; Yu, Yuanyuan
Conformation-specific Antibody Deciphers K27-linked Ubiquitination in Chaperone-Mediated Proteostasis Journal Article
In: bioRxiv, 2025, ISSN: 2692-8205.
@article{pmid41446272,
title = {Conformation-specific Antibody Deciphers K27-linked Ubiquitination in Chaperone-Mediated Proteostasis},
author = {Chengxiao Han and Yicheng Weng and Qingyun Zheng and Qian Qu and Satchal K Erramilli and Zhen Su and Yujuan Duan and Yunxi Han and Xiaoguo Zhai and Jingxian Li and Anthony A Kossiakoff and Man Pan and Minglei Zhao and Lei Liu and Yuanyuan Yu},
doi = {10.64898/2025.12.18.695067},
issn = {2692-8205},
year = {2025},
date = {2025-12-01},
urldate = {2025-12-01},
journal = {bioRxiv},
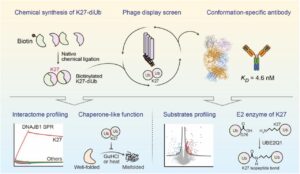
abstract = {Lysine 27 (K27)-linked polyubiquitination plays critical yet incompletely defined roles in proteostasis, innate immunity, and disease progression; however, investigations into this process have long been hindered by its extremely low abundance and the lack of conformation-specific enrichment tools. Herein, we describe the development of a long-sought conformation-specific antibody, K27-IgG, which can selectively recognize-among all ubiquitin chain types-the unique architecture of K27-linked polyubiquitin (K27-polyUb) characterized by a distinct buried K27-isopeptide bond, with high affinity (KD = 4.66 nM). This antibody was derived from synthetic antibodies initially generated via phage display, using chemically synthesized K27-linked diubiquitin (K27-diUb) as the antigen. High-resolution co-crystal structures uncovered the unique K27-diUb interface targeted by these sAbs. Subsequent reformatting of these sAbs into a full-length human immunoglobulin G (IgG) scaffold yielded K27-IgG, notably exhibiting markedly enhanced affinity without compromising selectivity. Using K27-IgG as a tool, we achieved sensitive detection and immunoprecipitation (IP) of endogenous K27-polyUb in cells, and delineated the intracellular interaction landscape of K27-polyUb through complementary proteomic approaches. Two key findings emerged: 1) The molecular chaperone DNAJB1 is a specific reader of K27-linked ubiquitin chains (but not other linkages) and that K27-polyUb chains themselves exhibit chaperone-like activity, suggesting a novel mechanism by which K27-polyUb regulates chaperone-mediated proteostasis; 2) The E2 enzyme UBE2Q1 assembles K27-diUb, identifying it as a potential writer for this ubiquitin chain topology. Collectively, this study establishes K27-IgG as a robust tool for deciphering the K27-linked ubiquitin code, thereby opening new avenues for investigating the biological functions of K27-linked polyubiquitination.nnHIGHLIGHTS: First K27-linkage conformation-specific antibody with nanomolar affinity overcomes a major barrier in the field.K27-IgG unlocks functional mapping of the K27 ubiquitin landscape under proteotoxic stress.Molecular chaperone DNAJB1 is a selective "reader" of K27-linked ubiquitin chains.K27 chains possess intrinsic chaperone activity, enabling protein refolding and suppressing aggregation.E2 enzyme UBE2Q1 is a "writer" that directly assembles K27-linked ubiquitin chains.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Zinkle, Allen P; Batista, Mariana Bunoro; Herrera, Carmen M; Erramilli, Satchal K; Kloss, Brian; Ashraf, Khuram U; Nosol, Kamil; Zhang, Guozhi; Cater, Rosemary J; Marty, Michael T; Kossiakoff, Anthony A; Trent, M Stephen; Nygaard, Rie; Stansfeld, Phillip J; Mancia, Filippo
Mechanistic basis of antimicrobial resistance mediated by the phosphoethanolamine transferase MCR-1 Journal Article
In: Nat Commun, vol. 16, no. 1, pp. 10516, 2025, ISSN: 2041-1723.
@article{pmid41298376,
title = {Mechanistic basis of antimicrobial resistance mediated by the phosphoethanolamine transferase MCR-1},
author = {Allen P Zinkle and Mariana Bunoro Batista and Carmen M Herrera and Satchal K Erramilli and Brian Kloss and Khuram U Ashraf and Kamil Nosol and Guozhi Zhang and Rosemary J Cater and Michael T Marty and Anthony A Kossiakoff and M Stephen Trent and Rie Nygaard and Phillip J Stansfeld and Filippo Mancia},
doi = {10.1038/s41467-025-65515-3},
issn = {2041-1723},
year = {2025},
date = {2025-11-01},
urldate = {2025-11-01},
journal = {Nat Commun},
volume = {16},
number = {1},
pages = {10516},
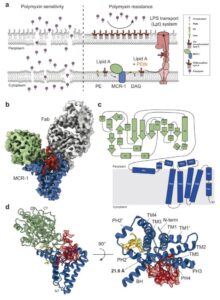
abstract = {Polymyxins are used to treat infections caused by multidrug-resistant Gram-negative bacteria. They are cationic peptides that target the negatively charged lipid A component of lipopolysaccharides, disrupting the outer membrane and lysing the cell. Polymyxin resistance is conferred by inner-membrane enzymes, such as phosphoethanolamine transferases, which add positively charged phosphoethanolamine to lipid A. Here, we present the structure of MCR-1, a plasmid-encoded phosphoethanolamine transferase, in its liganded form. The phosphatidylethanolamine donor substrate is bound near the active site in the periplasmic domain, and lipid A is bound over 20 Å away, within the transmembrane region. Integrating structural, biochemical, and drug-resistance data with computational analyses, we propose a two-state model in which the periplasmic domain rotates to bring the active site to lipid A, near the preferential phosphate modification site for MCR-1. This enzymatic mechanism may be generally applicable to other phosphoform transferases with large, globular soluble domains.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Romane, Ksenija; Peteani, Giulia; Mukherjee, Somnath; Kowal, Julia; Rossi, Lorenzo; Hou, Jingkai; Kossiakoff, Anthony A; Lemmin, Thomas; Locher, Kaspar P
Structural basis of drug recognition by human MATE1 transporter Journal Article
In: Nat Commun, vol. 16, no. 1, pp. 9444, 2025, ISSN: 2041-1723.
@article{pmid41145429,
title = {Structural basis of drug recognition by human MATE1 transporter},
author = {Ksenija Romane and Giulia Peteani and Somnath Mukherjee and Julia Kowal and Lorenzo Rossi and Jingkai Hou and Anthony A Kossiakoff and Thomas Lemmin and Kaspar P Locher},
doi = {10.1038/s41467-025-64490-z},
issn = {2041-1723},
year = {2025},
date = {2025-10-28},
urldate = {2025-10-01},
journal = {Nat Commun},
volume = {16},
number = {1},
pages = {9444},
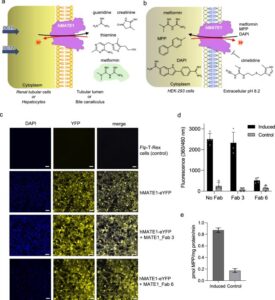
abstract = {Human MATE1 (multidrug and toxin extrusion protein 1) is highly expressed in the kidney and liver, where it mediates the final step in the excretion of a broad range of cationic drugs, including the antidiabetic drug metformin, into the urine and bile. This transport process is essential for drug clearance and also affects therapeutic efficacy. To understand the molecular basis of drug recognition by hMATE1, we determined cryo-electron microscopy structures of the transporter in complex with the substrates 1-methyl-4-phenylpyridinium (MPP) and metformin and with the inhibitor cimetidine. The structures reveal a shared binding site located in a negatively charged pocket in the C-lobe of the protein. We functionally validated key interactions using radioactivity-based cellular uptake assays using hMATE1 mutants. Molecular dynamics simulations provide insights into the different binding modes and dynamic behaviour of the ligands within the pocket. Collectively, these findings define the structural basis of hMATE1 substrate specificity and shed light on its role in drug transport and drug-drug interactions.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Mohona, Surovi; Shakya, Anil K; Singh, Suruchi; Kearns, Fiona L; Jemison, Kezia; Erramilli, Satchal; Dey, Debajit; Qing, Enya; Jennings, Benjamin C; Doray, Balraj; Kossiakoff, Anthony A; Amaro, Rommie E; Klose, Thomas; Gallagher, Tom; Hasan, S Saif
An unconventional HxD motif orchestrates coatomer-dependent coronavirus morphogenesis Journal Article
In: bioRxiv, 2025, ISSN: 2692-8205.
@article{pmid41279991,
title = {An unconventional HxD motif orchestrates coatomer-dependent coronavirus morphogenesis},
author = {Surovi Mohona and Anil K Shakya and Suruchi Singh and Fiona L Kearns and Kezia Jemison and Satchal Erramilli and Debajit Dey and Enya Qing and Benjamin C Jennings and Balraj Doray and Anthony A Kossiakoff and Rommie E Amaro and Thomas Klose and Tom Gallagher and S Saif Hasan},
doi = {10.1101/2025.10.16.682669},
issn = {2692-8205},
year = {2025},
date = {2025-10-17},
urldate = {2025-10-01},
journal = {bioRxiv},
abstract = {Assembly of infectious coronaviruses requires spike (S) protein trafficking by host coatomer, typically via a dibasic signal in the S cytoplasmic tail. However, the human embecoviruses HKU1 and OC43, as well as the model virus MHV, lack this motif. Here we identify a conserved His-x-Asp (HxD) sequence that functions as an unconventional coatomer-binding signal. Structural and biochemical analyses show that the MHV HxD motif engages coatomer subunits through distinct conformations, while cellular imaging demonstrates its role in directing S to assembly sites with the viral M-protein. Disruption of HxD-coatomer interactions impairs S incorporation and provokes compensatory viral adaptations, including emergence of a canonical dibasic motif or mutations in M-protein. Electron microscopy further reveals profound alterations in virion surface architecture. These findings uncover HxD as a previously unrecognized coatomer-targeting motif, highlighting an unexpected flexibility in coronavirus assembly pathways and broadening understanding of the cellular machinery that shapes coronavirus morphogenesis.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
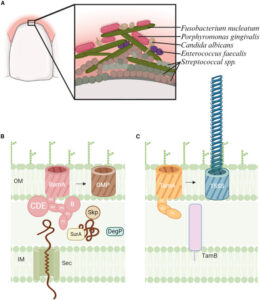
Cottom, Claire Overly; Heinz, Eva; Erramilli, Satchal; Kossiakoff, Anthony; Slade, Daniel J; Noinaj, Nicholas
Characterization of the OMP biogenesis machinery in Fusobacterium nucleatum Journal Article
In: Structure, vol. 33, iss. 33, no. 11, pp. 1878–1892.e5, 2025, ISSN: 1878-4186.
@article{pmid40897170,
title = {Characterization of the OMP biogenesis machinery in Fusobacterium nucleatum},
author = {Claire Overly Cottom and Eva Heinz and Satchal Erramilli and Anthony Kossiakoff and Daniel J Slade and Nicholas Noinaj},
doi = {10.1016/j.str.2025.08.008},
issn = {1878-4186},
year = {2025},
date = {2025-09-01},
urldate = {2025-09-01},
journal = {Structure},
volume = {33},
number = {11},
issue = {33},
pages = {1878--1892.e5},
abstract = {F. nucleatum is a Gram-negative bacteria that causes oral infections and is linked to colorectal cancer. Pathogenicity relies on a type of β-barrel outer membrane protein (OMP) called an autotransporter. The biogenesis of OMPs is typically mediated by the barrel assembly machinery (BAM) complex. In this study, we investigate the evolution, composition, and structure of the OMP biogenesis machinery in F. nucleatum. Our bioinformatics and proteomics analyses indicate that OMP biogenesis in F. nucleatum is mediated solely by the core component BamA. The structure of FnBamA highlights distinct features, including four POTRA domains and a C-terminal 16-stranded β-barrel domain observed as an inverted dimer. FnBamA represents the original composition of the assembly machinery, and a duplication event that resulted in BamA and TamA occurred after the split of other lineages, including the Proteobacteria, from the Fusobacteria. FnBamA, therefore, likely serves a singular role in the biogenesis of all OMPs.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
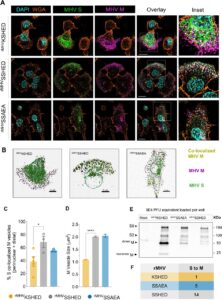
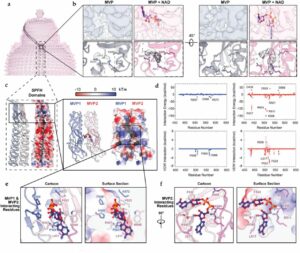
Lodwick, Jane E; Shen, Rong; Erramilli, Satchal; Xie, Yuan; Roganowicz, Karolina; Kossiakoff, Anthony A; Zhao, Minglei
Structural Insights into the Roles of PARP4 and NAD in the Human Vault Cage Journal Article
In: Nature Communications, iss. 16, no. 6724, 2025, ISSN: 2692-8205.
@article{pmid38979142,
title = {Structural Insights into the Roles of PARP4 and NAD in the Human Vault Cage},
author = {Jane E Lodwick and Rong Shen and Satchal Erramilli and Yuan Xie and Karolina Roganowicz and Anthony A Kossiakoff and Minglei Zhao},
doi = {10.1038/s41467-025-61981-x},
issn = {2692-8205},
year = {2025},
date = {2025-07-21},
urldate = {2024-06-01},
journal = {Nature Communications},
number = {6724},
issue = {16},
abstract = {Vault is a massive ribonucleoprotein complex found across Eukaryota. The major vault protein (MVP) oligomerizes into an ovular cage, which contains several minor vault components (MVCs) and is thought to transport transiently bound "cargo" molecules. Vertebrate vaults house a poly (ADP-ribose) polymerase (known as PARP4 in humans), which is the only MVC with known enzymatic activity. Despite being discovered decades ago, the molecular basis for PARP4's interaction with MVP remains unclear. In this study, we determined the structure of the human vault cage in complex with PARP4 and its enzymatic substrate NAD . The structures reveal atomic-level details of the protein-binding interface, as well as unexpected NAD -binding pockets within the interior of the vault cage. In addition, proteomics data show that human vaults purified from wild-type and PARP4-depleted cells interact with distinct subsets of proteins. Our results thereby support a model in which PARP4's specific incorporation into the vault cage helps to regulate vault's selection of cargo and its subcellular localization. Further, PARP4's proximity to MVP's NAD -binding sites could support its enzymatic function within the vault.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
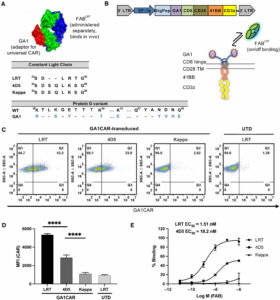
Arina, Ainhoa; Arauz, Edwin; Masoumi, Elham; Warzecha, Karolina W; Sääf, Annika; Widło, Łukasz; Slezak, Tomasz; Zieminska, Aleksandra; Dudek, Karolina; Schaefer, Zachary P; Lecka, Maria; Usatyuk, Svitlana; Weichselbaum, Ralph R; Kossiakoff, Anthony A
A universal chimeric antigen receptor (CAR)-fragment antibody binder (FAB) split system for cancer immunotherapy Journal Article
In: Sci Adv, vol. 11, no. 27, pp. eadv4937, 2025, ISSN: 2375-2548.
@article{pmid40614208,
title = {A universal chimeric antigen receptor (CAR)-fragment antibody binder (FAB) split system for cancer immunotherapy},
author = {Ainhoa Arina and Edwin Arauz and Elham Masoumi and Karolina W Warzecha and Annika Sääf and Łukasz Widło and Tomasz Slezak and Aleksandra Zieminska and Karolina Dudek and Zachary P Schaefer and Maria Lecka and Svitlana Usatyuk and Ralph R Weichselbaum and Anthony A Kossiakoff},
doi = {10.1126/sciadv.adv4937},
issn = {2375-2548},
year = {2025},
date = {2025-07-01},
urldate = {2025-07-01},
journal = {Sci Adv},
volume = {11},
number = {27},
pages = {eadv4937},
abstract = {Chimeric antigen receptor (CAR) T cell therapy has shown extraordinary results in treating hematological cancer but faces challenges like antigen loss, toxicity, and complex manufacturing. Universal and modular CAR constructs offer improved flexibility, safety, and cost-effectiveness over conventional CAR constructs. We present a CAR-fragment antibody binder (Fab) platform on the basis of an engineered protein G variant (GA1) and Fab scaffolds. Expression of GA1CAR on human CD8 T cells leads to antigen recognition and T cell effector function that can be modulated according to the affinity of the CAR for the Fab and of the Fab for the target. GA1CAR T cells can recognize multiple Fab-antigen pairs on breast and ovarian cancer cell lines. Adoptively transferred GA1CAR T cells control tumors in breast cancer xenograft models, and their targeting can be quickly redirected using different Fabs. This versatile "plug-and-play" CAR T platform has potential for application in personalized therapy, preventing antigen loss variant escape, decreasing toxicity, and increasing access.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Ogbu, Chinemerem P; de Las Alas, Mason; Mandriota, Alexandria M; Liu, Xiangdong; Kapoor, Srajan; Choudhury, Jagrity; Ruma, Yasmeen N; Goodman, Michael C; Sanders, Charles R; Gonen, Tamir; Kossiakoff, Anthony A; Duffey, Michael E; Vecchio, Alex J
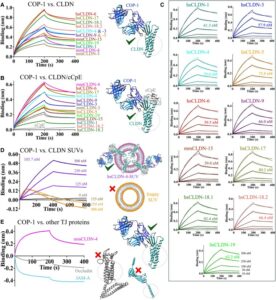
Biophysical basis of tight junction barrier modulation by a pan-claudin-binding molecule Journal Article
In: PNAS Nexus, vol. 4, no. 6, pp. pgaf189, 2025, ISSN: 2752-6542.
@article{pmid40575703,
title = {Biophysical basis of tight junction barrier modulation by a pan-claudin-binding molecule},
author = {Chinemerem P Ogbu and Mason de Las Alas and Alexandria M Mandriota and Xiangdong Liu and Srajan Kapoor and Jagrity Choudhury and Yasmeen N Ruma and Michael C Goodman and Charles R Sanders and Tamir Gonen and Anthony A Kossiakoff and Michael E Duffey and Alex J Vecchio},
doi = {10.1093/pnasnexus/pgaf189},
issn = {2752-6542},
year = {2025},
date = {2025-06-01},
urldate = {2025-06-01},
journal = {PNAS Nexus},
volume = {4},
number = {6},
pages = {pgaf189},
abstract = {Claudins are a 27-member family of membrane proteins that form and fortify specialized cell contacts in endothelium and epithelium called tight junctions. Tight junctions restrict paracellular transport through tissues by forming molecular barriers between cells. Claudin-binding molecules thus hold promise for modulating tight junction permeability to deliver drugs or as therapeutics to treat tight junction-linked disease. The development of claudin-binding molecules, however, is hindered by their physicochemical intractability and small targetable surfaces. Here, we determine that a synthetic antibody fragment (sFab) that we developed binds with nanomolar affinity directly to 10 claudin subtypes and other distantly related claudin family members but not to other tight junction-localized membrane proteins. It does so by targeting the extracellular surfaces of claudins, which we verify by applying this sFab to a model intestinal epithelium and observe that it opens paracellular barriers comparable to a known, but application limited, tight junction modulating protein. This pan-claudin-binding molecule holds potential for both basic and translational applications as it is a probe of claudin and tight junction structure in vitro and in vivo and a tool to modulate the permeability of tight junctions broadly across tissue barriers.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
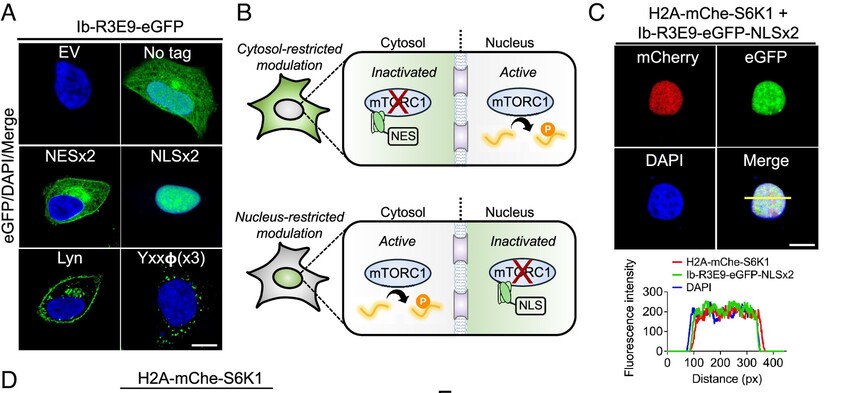
O'Leary, Kelly M; Slezak, Tomasz; Kossiakoff, Anthony A
Conformation-specific synthetic intrabodies modulate mTOR signaling with subcellular spatial resolution Journal Article
In: Proc Natl Acad Sci U S A, vol. 122, no. 24, pp. e2424679122, 2025, ISSN: 1091-6490.
@article{pmid40489625,
title = {Conformation-specific synthetic intrabodies modulate mTOR signaling with subcellular spatial resolution},
author = {Kelly M O'Leary and Tomasz Slezak and Anthony A Kossiakoff},
doi = {10.1073/pnas.2424679122},
issn = {1091-6490},
year = {2025},
date = {2025-06-01},
urldate = {2025-06-01},
journal = {Proc Natl Acad Sci U S A},
volume = {122},
number = {24},
pages = {e2424679122},
abstract = {Subcellular compartmentalization is integral to the spatial regulation of mechanistic target of rapamycin (mTOR) signaling. However, the biological outputs associated with location-specific mTOR signaling events are poorly understood and challenging to decouple. Here, we engineered synthetic intracellular antibodies (intrabodies) that are capable of modulating mTOR signaling with genetically programmable spatial resolution. Epitope-directed phage display was exploited to generate high affinity synthetic antibody fragments (Fabs) against the FKBP12-Rapamycin binding site of mTOR (mTOR). We determined high-resolution crystal structures of two unique Fabs that discriminate distinct conformational states of mTOR through recognition of its substrate recruitment interface. By leveraging these conformation-specific binders as intracellular probes, we uncovered the structural basis for an allosteric mechanism governing mTOR complex 1 (mTORC1) stability mediated by subtle structural adjustments within mTOR. Furthermore, our results demonstrated that synthetic binders emulate natural substrates by employing divergent yet complementary hydrophobic residues at defined positions, underscoring the broad molecular recognition capability of mTOR. Intracellular signaling studies showed differential time-dependent inhibition of S6 kinase 1 and Akt phosphorylation by genetically encoded intrabodies, thus supporting a mechanism of inhibition analogous to the natural product rapamycin. Finally, we implemented a feasible approach to selectively modulate mTOR signaling in the nucleus through spatially programmed intrabody expression. These findings establish intrabodies as versatile tools for dissecting the conformational regulation of mTORC1 and should be useful to explore how location-specific mTOR signaling influences disease progression.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Chen, Qiuyan; Schafer, Christopher T; Mukherjee, Somnath; Wang, Kai; Gustavsson, Martin; Fuller, James R; Tepper, Katelyn; Lamme, Thomas D; Aydin, Yasmin; Agrawal, Parth; Terashi, Genki; Yao, Xin-Qiu; Kihara, Daisuke; Kossiakoff, Anthony A; Handel, Tracy M; Tesmer, John J G
Effect of phosphorylation barcodes on arrestin binding to a chemokine receptor Journal Article
In: Nature, 2025, ISSN: 1476-4687.
@article{pmid40399676,
title = {Effect of phosphorylation barcodes on arrestin binding to a chemokine receptor},
author = {Qiuyan Chen and Christopher T Schafer and Somnath Mukherjee and Kai Wang and Martin Gustavsson and James R Fuller and Katelyn Tepper and Thomas D Lamme and Yasmin Aydin and Parth Agrawal and Genki Terashi and Xin-Qiu Yao and Daisuke Kihara and Anthony A Kossiakoff and Tracy M Handel and John J G Tesmer},
doi = {10.1038/s41586-025-09024-9},
issn = {1476-4687},
year = {2025},
date = {2025-05-01},
urldate = {2025-05-01},
journal = {Nature},
abstract = {Unique phosphorylation 'barcodes' installed in different regions of an active seven-transmembrane receptor by different G-protein-coupled receptor (GPCR) kinases (GRKs) have been proposed to promote distinct cellular outcomes, but it is unclear whether or how arrestins differentially engage these barcodes. Here, to address this, we developed an antigen-binding fragment (Fab7) that recognizes both active arrestin2 (β-arrestin1) and arrestin3 (β-arrestin2) without interacting with bound receptor polypeptides. We used Fab7 to determine the structures of both arrestins in complex with atypical chemokine receptor 3 (ACKR3) phosphorylated in different regions of its C-terminal tail by either GRK2 or GRK5 (ref. ). The GRK2-phosphorylated ACKR3 resulted in more heterogeneous 'tail-mode' assemblies, whereas phosphorylation by GRK5 resulted in more rigid 'ACKR3-adjacent' assemblies. Unexpectedly, the finger loops of both arrestins engaged the micelle surface rather than the receptor intracellular pocket, with arrestin3 being more dynamic, partly because of its lack of a membrane-anchoring motif. Thus, both the region of the barcode and the arrestin isoform involved can alter the structure and dynamics of GPCR-arrestin complexes, providing a possible mechanistic basis for unique downstream cellular effects, such as the efficiency of chemokine scavenging and the robustness of arrestin binding in ACKR3.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

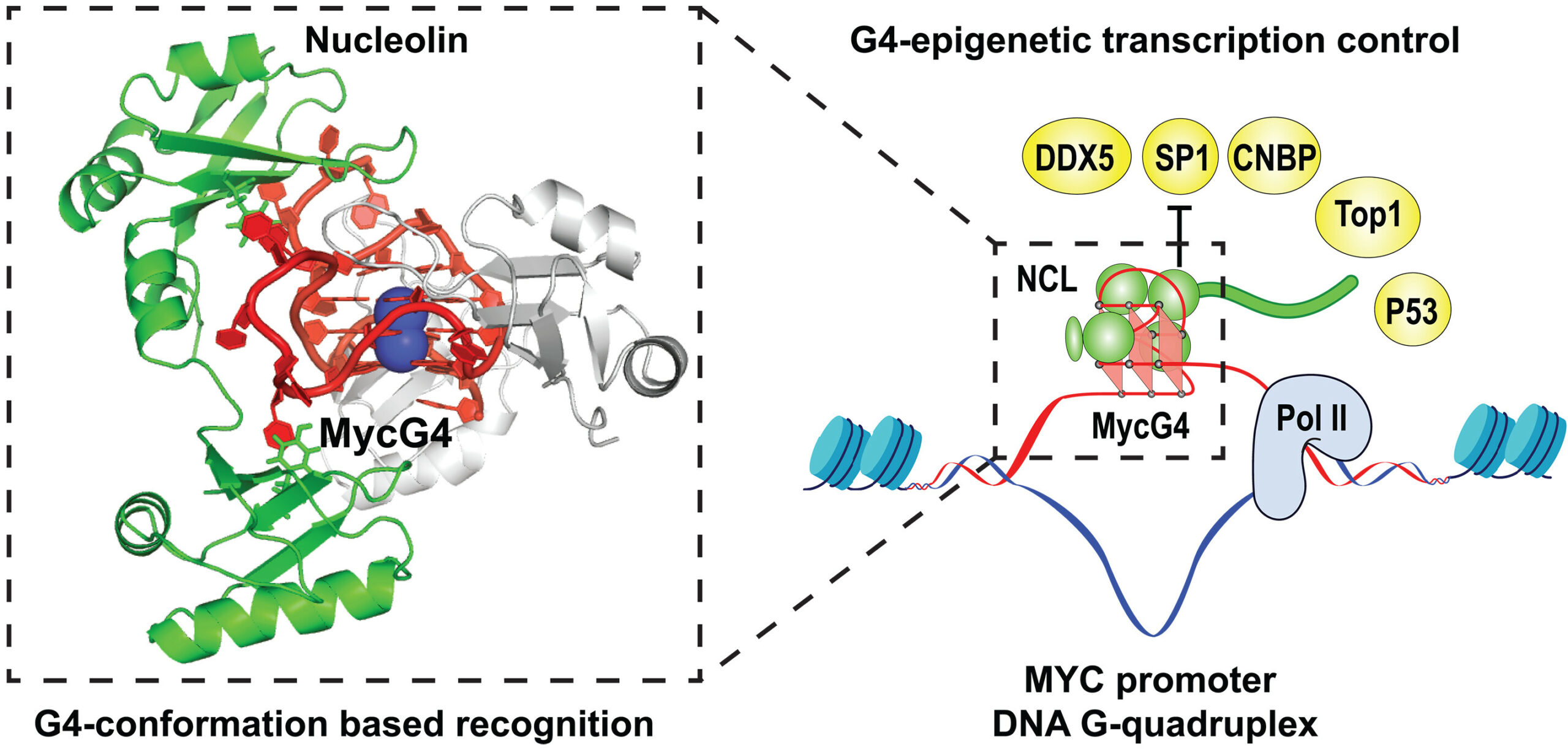
Chen, Luying; Dickerhoff, Jonathan; Zheng, Ke-wei; Erramilli, Satchal; Feng, Hanqiao; Wu, Guanhui; Onel, Buket; Chen, Yuwei; Wang, Kai-Bo; Carver, Megan; Lin, Clement; Sakai, Saburo; Wan, Jun; Vinson, Charles; Hurley, Laurence; Kossiakoff, Anthony A.; Deng, Nanjie; Bai, Yawen; Noinaj, Nicholas; Yang, Danzhou
Structural basis for nucleolin recognition of MYC promoter G-quadruplex Journal Article
In: Science, vol. 388, no. 6744, 2025, ISSN: 1095-9203.
@article{Chen2025,
title = {Structural basis for nucleolin recognition of MYC promoter G-quadruplex},
author = {Luying Chen and Jonathan Dickerhoff and Ke-wei Zheng and Satchal Erramilli and Hanqiao Feng and Guanhui Wu and Buket Onel and Yuwei Chen and Kai-Bo Wang and Megan Carver and Clement Lin and Saburo Sakai and Jun Wan and Charles Vinson and Laurence Hurley and Anthony A. Kossiakoff and Nanjie Deng and Yawen Bai and Nicholas Noinaj and Danzhou Yang},
doi = {10.1126/science.adr1752},
issn = {1095-9203},
year = {2025},
date = {2025-04-18},
urldate = {2025-04-18},
journal = {Science},
volume = {388},
number = {6744},
publisher = {American Association for the Advancement of Science (AAAS)},
abstract = {The MYC oncogene promoter G-quadruplex (MycG4) regulates transcription and is a prevalent G4 locus in immortal cells. Nucleolin, a major MycG4-binding protein, exhibits greater affinity for MycG4 than for nucleolin recognition element (NRE) RNA. Nucleolin’s four RNA binding domains (RBDs) are essential for high-affinity MycG4 binding. We present the 2.6-angstrom crystal structure of the nucleolin-MycG4 complex, revealing a folded parallel three-tetrad G-quadruplex with two coordinating potassium ions (K+), interacting with RBD1, RBD2, and Linker12 through its 6–nucleotide (nt) central loop and 5′ flanking region. RBD3 and RBD4 bind MycG4’s 1-nt loops as demonstrated by nuclear magnetic resonance (NMR). Cleavage under targets and tagmentation sequencing confirmed nucleolin’s binding to MycG4 in cells. Our results revealed a G4 conformation-based recognition by a regulating protein through multivalent interactions, suggesting that G4s are nucleolin’s primary cellular substrates, indicating G4 epigenetic transcriptional regulation and helping G4-targeted drug discovery.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
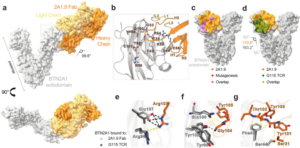
Ramesh, Amrita; Roy, Sobhan; Slezak, Tomasz; Fuller, James; Graves, Hortencia; Mamedov, Murad R.; Marson, Alexander; Kossiakoff, Anthony A.; Adams, Erin J.
Mapping the extracellular molecular architecture of the pAg-signaling complex with α-Butyrophilin antibodies Journal Article
In: Sci Rep, vol. 15, no. 1, 2025, ISSN: 2045-2322.
@article{Ramesh2025,
title = {Mapping the extracellular molecular architecture of the pAg-signaling complex with α-Butyrophilin antibodies},
author = {Amrita Ramesh and Sobhan Roy and Tomasz Slezak and James Fuller and Hortencia Graves and Murad R. Mamedov and Alexander Marson and Anthony A. Kossiakoff and Erin J. Adams},
doi = {10.1038/s41598-025-94347-w},
issn = {2045-2322},
year = {2025},
date = {2025-04-09},
urldate = {2025-12-00},
journal = {Sci Rep},
volume = {15},
number = {1},
publisher = {Springer Science and Business Media LLC},
abstract = {Target cells trigger Vγ9Vδ2 T cell activation by signaling the intracellular accumulation of phospho-antigen metabolites (pAgs) through Butyrophilin (BTN)-3A1 and BTN2A1 to the Vγ9Vδ2 T cell receptor (TCR). An incomplete understanding of the molecular dynamics in this signaling complex hampers Vγ9Vδ2 T cell immunotherapeutic efficacy. A panel of engineered α-BTN3A1 and α-BTN2A1 antibody (mAb) reagents was used to probe the roles of BTN3A1 and BTN2A1 in pAg signaling. Modified α-BTN3A1 mAbs with increased inter-Fab distances establish that tight clustering of BTN3A1 is not necessary to stimulate Vγ9Vδ2 T cell activation, and that antagonism may occur through occlusion of a critical binding interaction between BTN3A1 and a yet unknown co-receptor. Finally, a panel of additional α-BTN2A1 antagonists utilize different biophysical mechanisms to compete with Vγ9Vδ2 TCRs for BTN2A1 binding. The complex structures of BTN2A1 ectodomain and Fabs from three antagonist mAbs provide molecular insights into BTN2A1 epitopes critical for pAg-signaling.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
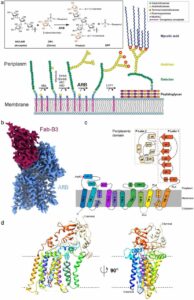
Liu, Yaqi; Brown, Chelsea M; Erramilli, Satchal; Su, Yi-Chia; Tseng, Po-Sen; Wang, Yu-Jen; Duong, Nam Ha; Tokarz, Piotr; Kloss, Brian; Han, Cheng-Ruei; Chen, Hung-Yu; Rodrigues, Jose; Archer, Margarida; Lowary, Todd L; Kossiakoff, Anthony A; Stansfeld, Phillip J; Nygaard, Rie; Mancia, Filippo
Structural insights into terminal arabinosylation biosynthesis of the mycobacterial cell wall arabinan Journal Article
In: Nature Communications, 2025, ISSN: 2692-8205.
@article{pmid39345558,
title = {Structural insights into terminal arabinosylation biosynthesis of the mycobacterial cell wall arabinan},
author = {Yaqi Liu and Chelsea M Brown and Satchal Erramilli and Yi-Chia Su and Po-Sen Tseng and Yu-Jen Wang and Nam Ha Duong and Piotr Tokarz and Brian Kloss and Cheng-Ruei Han and Hung-Yu Chen and Jose Rodrigues and Margarida Archer and Todd L Lowary and Anthony A Kossiakoff and Phillip J Stansfeld and Rie Nygaard and Filippo Mancia},
doi = {10.1038/s41467-025-58196-5},
issn = {2692-8205},
year = {2025},
date = {2025-03-14},
urldate = {2024-09-01},
journal = {Nature Communications},
abstract = {The emergence of drug-resistant strains exacerbates the global challenge of tuberculosis caused by Mycobacterium tuberculosis (Mtb). Central to the pathogenicity of Mtb is its complex cell envelope, which serves as a barrier against both immune system and pharmacological attacks. Two key components of this envelope, arabinogalactan (AG) and lipoarabinomannan (LAM) are complex polysaccharides that contain integral arabinan domains important for cell wall structural and functional integrity. The arabinofuranosyltransferase AftB terminates the synthesis of these arabinan domains by catalyzing the addition of the addition of β-(1→2)-linked terminal arabinofuranose residues. Here, we present the cryo-EM structures of Mycobacterium chubuense AftB in its apo and donor substrate analog-bound form, determined to 2.9 Å and 3.4 Å resolution, respectively. Our structures reveal that AftB has a GT-C fold transmembrane (TM) domain comprised of eleven TM helices and a periplasmic cap domain. AftB has an irregular tube-shaped cavity that bridges the two proposed substrate binding sites. By integrating structural analysis, biochemical assays, and molecular dynamics simulations, we elucidate the molecular basis of the reaction mechanism of AftB and propose a model for catalysis.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
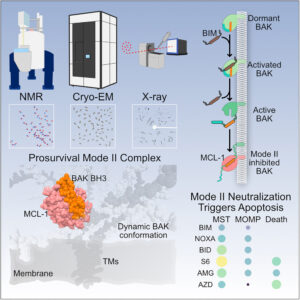
Srivastava, Shagun; Sekar, Giridhar; Ojoawo, Adedolapo; Aggarwal, Anup; Ferreira, Elisabeth; Uchikawa, Emiko; Yang, Meek; Grace, Christy R; Dey, Raja; Lin, Yi-Lun; Guibao, Cristina D; Jayaraman, Seetharaman; Mukherjee, Somnath; Kossiakoff, Anthony A; Dong, Bin; Myasnikov, Alexander; Moldoveanu, Tudor
Structural basis of BAK sequestration by MCL-1 in apoptosis Journal Article
In: Mol Cell, 2025, ISSN: 1097-4164.
@article{pmid40187349,
title = {Structural basis of BAK sequestration by MCL-1 in apoptosis},
author = {Shagun Srivastava and Giridhar Sekar and Adedolapo Ojoawo and Anup Aggarwal and Elisabeth Ferreira and Emiko Uchikawa and Meek Yang and Christy R Grace and Raja Dey and Yi-Lun Lin and Cristina D Guibao and Seetharaman Jayaraman and Somnath Mukherjee and Anthony A Kossiakoff and Bin Dong and Alexander Myasnikov and Tudor Moldoveanu},
doi = {10.1016/j.molcel.2025.03.013},
issn = {1097-4164},
year = {2025},
date = {2025-03-01},
urldate = {2025-03-01},
journal = {Mol Cell},
abstract = {Apoptosis controls cell fate, ensuring tissue homeostasis and promoting disease when dysregulated. The rate-limiting step in apoptosis is mitochondrial poration by the effector B cell lymphoma 2 (BCL-2) family proteins BAK and BAX, which are activated by initiator BCL-2 homology 3 (BH3)-only proteins (e.g., BIM) and inhibited by guardian BCL-2 family proteins (e.g., MCL-1). We integrated structural, biochemical, and pharmacological approaches to characterize the human prosurvival MCL-1:BAK complex assembled from their BCL-2 globular core domains. We reveal a canonical interaction with BAK BH3 bound to the hydrophobic groove of MCL-1 and disordered and highly dynamic BAK regions outside the complex interface. We predict similar conformations of activated effectors in complex with other guardians or effectors. The MCL-1:BAK complex is a major cancer drug target. We show that MCL-1 inhibitors are inefficient in neutralizing the MCL-1:BAK complex, requiring high doses to initiate apoptosis. Our study underscores the need to design superior clinical candidate MCL-1 inhibitors.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
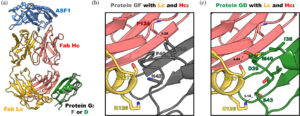
Slezak, Tomasz; O'Leary, Kelly M; Li, Jinyang; Rohaim, Ahmed; Davydova, Elena K; Kossiakoff, Anthony A
Engineered protein G variants for multifunctional antibody-based assemblies Journal Article
In: Protein Sci, vol. 34, no. 2, pp. e70019, 2025, ISSN: 1469-896X.
@article{pmid39865354,
title = {Engineered protein G variants for multifunctional antibody-based assemblies},
author = {Tomasz Slezak and Kelly M O'Leary and Jinyang Li and Ahmed Rohaim and Elena K Davydova and Anthony A Kossiakoff},
doi = {10.1002/pro.70019},
issn = {1469-896X},
year = {2025},
date = {2025-02-01},
urldate = {2025-02-01},
journal = {Protein Sci},
volume = {34},
number = {2},
pages = {e70019},
abstract = {We have developed a portfolio of antibody-based modules that can be prefabricated as standalone units and snapped together in plug-and-play fashion to create uniquely powerful multifunctional assemblies. The basic building blocks are derived from multiple pairs of native and modified Fab scaffolds and protein G (PG) variants engineered by phage display to introduce high pair-wise specificity. The variety of possible Fab-PG pairings provides a highly orthogonal system that can be exploited to perform challenging cell biology operations in a straightforward manner. The simplest manifestation allows multiplexed antigen detection using PG variants fused to fluorescently labeled SNAP-tags. Moreover, Fabs can be readily attached to a PG-Fc dimer module which acts as the core unit to produce plug-and-play IgG-like assemblies, and the utility can be further expanded to produce bispecific analogs using the "knobs into holes" strategy. These core PG-Fc dimer modules can be made and stored in bulk to produce off-the-shelf customized IgG entities in minutes, not days or weeks by just adding a Fab with the desired antigen specificity. In another application, the bispecific modalities form the building block for fabricating potent bispecific T-cell engagers (BiTEs), demonstrating their efficacy in cancer cell-killing assays. Additionally, the system can be adapted to include commercial antibodies as building blocks, greatly increasing the target space. Crystal structure analysis reveals that a few strategically positioned interactions engender the specificity between the Fab-PG variant pairs, requiring minimal changes to match the scaffolds for different possible combinations. This plug-and-play platform offers a user-friendly and versatile approach to enhance the functionality of antibody-based reagents in cell biology research.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
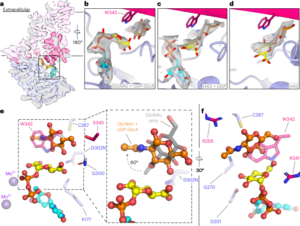
Górniak, Ireneusz; Stephens, Zachery; Erramilli, Satchal K; Gawda, Tomasz; Kossiakoff, Anthony A; Zimmer, Jochen
Structural insights into translocation and tailored synthesis of hyaluronan Journal Article
In: Nat Struct Mol Biol, vol. 32, no. 1, pp. 161–171, 2025, ISSN: 1545-9985.
@article{pmid39322765,
title = {Structural insights into translocation and tailored synthesis of hyaluronan},
author = {Ireneusz Górniak and Zachery Stephens and Satchal K Erramilli and Tomasz Gawda and Anthony A Kossiakoff and Jochen Zimmer},
doi = {10.1038/s41594-024-01389-1},
issn = {1545-9985},
year = {2025},
date = {2025-01-01},
urldate = {2025-01-01},
journal = {Nat Struct Mol Biol},
volume = {32},
number = {1},
pages = {161--171},
abstract = {Hyaluronan (HA) is an essential component of the vertebrate extracellular matrix. It is a heteropolysaccharide of N-acetylglucosamine (GlcNAc) and glucuronic acid (GlcA) reaching several megadaltons in healthy tissues. HA is synthesized and translocated in a coupled reaction by HA synthase (HAS). Here, structural snapshots of HAS provide insights into HA biosynthesis, from substrate recognition to HA elongation and translocation. We monitor the extension of a GlcNAc primer with GlcA, reveal the coordination of the uridine diphosphate product by a conserved gating loop and capture the opening of a translocation channel to coordinate a translocating HA polymer. Furthermore, we identify channel-lining residues that modulate HA product lengths. Integrating structural and biochemical analyses suggests an avenue for polysaccharide engineering based on finely tuned enzymatic activity and HA coordination.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
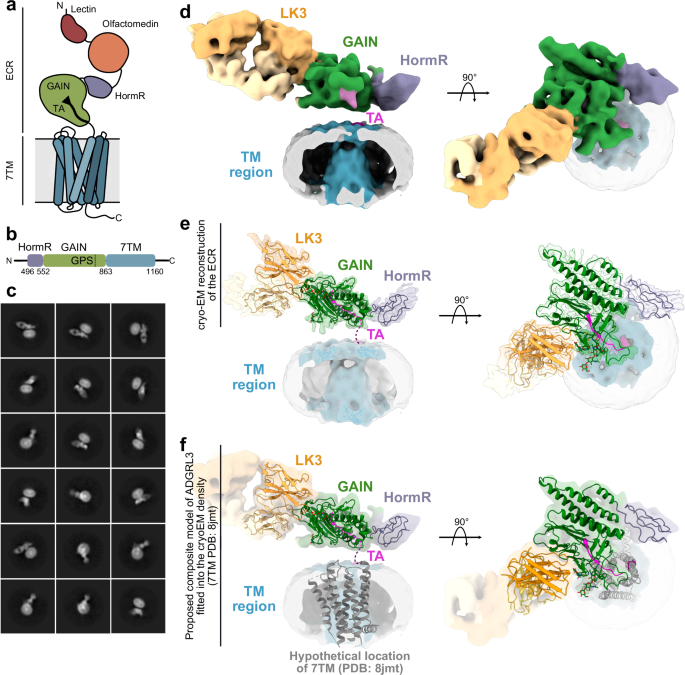
Kordon, Szymon P; Cechova, Kristina; Bandekar, Sumit J; Leon, Katherine; Dutka, Przemysław; Siffer, Gracie; Kossiakoff, Anthony A; Vafabakhsh, Reza; Araç, Demet
Conformational coupling between extracellular and transmembrane domains modulates holo-adhesion GPCR function Journal Article
In: Nat Commun, vol. 15, no. 1, pp. 10545, 2024, ISSN: 2041-1723.
@article{pmid39627215,
title = {Conformational coupling between extracellular and transmembrane domains modulates holo-adhesion GPCR function},
author = {Szymon P Kordon and Kristina Cechova and Sumit J Bandekar and Katherine Leon and Przemysław Dutka and Gracie Siffer and Anthony A Kossiakoff and Reza Vafabakhsh and Demet Araç},
doi = {10.1038/s41467-024-54836-4},
issn = {2041-1723},
year = {2024},
date = {2024-12-01},
urldate = {2024-12-01},
journal = {Nat Commun},
volume = {15},
number = {1},
pages = {10545},
abstract = {Adhesion G Protein-Coupled Receptors (aGPCRs) are key cell-adhesion molecules involved in numerous physiological functions. aGPCRs have large multi-domain extracellular regions (ECRs) containing a conserved GAIN domain that precedes their seven-pass transmembrane domain (7TM). Ligand binding and mechanical force applied on the ECR regulate receptor function. However, how the ECR communicates with the 7TM remains elusive, because the relative orientation and dynamics of the ECR and 7TM within a holoreceptor is unclear. Here, we describe the cryo-EM reconstruction of an aGPCR, Latrophilin3/ADGRL3, and reveal that the GAIN domain adopts a parallel orientation to the transmembrane region and has constrained movement. Single-molecule FRET experiments unveil three slow-exchanging FRET states of the ECR relative to the transmembrane region within the holoreceptor. GAIN-targeted antibodies, and cancer-associated mutations at the GAIN-7TM interface, alter FRET states, cryo-EM conformations, and receptor signaling. Altogether, this data demonstrates conformational and functional coupling between the ECR and 7TM, suggesting an ECR-mediated mechanism for aGPCR activation.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
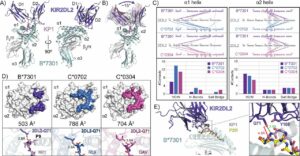
Ross, Philipp; Hilton, Hugo G; Lodwick, Jane; Slezak, Tomasz; Guethlein, Lisbeth A; McMurtrey, Curtis P; Han, Alex S; Nielsen, Morten; Yong, Daniel; Dulberger, Charles L; Nolan, Kristof T; Roy, Sobhan; Castro, Caitlin D; Hildebrand, William H; Zhao, Minglei; Kossiakoff, Anthony; Parham, Peter; Adams, Erin J
Molecular characterization of the archaic HLA-B*73:01 allele reveals presentation of a unique peptidome and skewed engagement by KIR2DL2 Journal Article
In: bioRxiv, 2024, ISSN: 2692-8205.
@article{pmid39651149,
title = {Molecular characterization of the archaic HLA-B*73:01 allele reveals presentation of a unique peptidome and skewed engagement by KIR2DL2},
author = {Philipp Ross and Hugo G Hilton and Jane Lodwick and Tomasz Slezak and Lisbeth A Guethlein and Curtis P McMurtrey and Alex S Han and Morten Nielsen and Daniel Yong and Charles L Dulberger and Kristof T Nolan and Sobhan Roy and Caitlin D Castro and William H Hildebrand and Minglei Zhao and Anthony Kossiakoff and Peter Parham and Erin J Adams},
doi = {10.1101/2024.11.25.625330},
issn = {2692-8205},
year = {2024},
date = {2024-11-01},
urldate = {2024-11-01},
journal = {bioRxiv},
abstract = {HLA class I alleles of archaic origin may have been retained in modern humans because they provide immunity against diseases to which archaic humans had evolved resistance. According to this model, archaic introgressed alleles were somehow distinct from those that evolved in African populations. Here we show that HLA-B*73:01, a rare allotype with putative archaic origins, has a relatively rare peptide binding motif with an unusually long-tailed peptide length distribution. We also find that HLA-B*73:01 combines a restricted and unique peptidome with high-cell surface expression, characteristics that make it well-suited to combat one or a number of closely-related pathogens. Furthermore, a crystal structure of HLA-B*73:01 in complex with KIR2DL2 highlights differences from previously solved structures with HLA-C molecules. These molecular characteristics distinguish HLA-B*73:01 from other HLA class I alleles previously investigated and may have provided early modern human migrants that inherited this allele with a selective advantage as they colonized Europe and Asia.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Rathnayake, Sewwandi S; Erramilli, Satchal K; Kossiakoff, Anthony A; Vecchio, Alex J
Cryo-EM structures of Clostridium perfringens enterotoxin bound to its human receptor, claudin-4 Journal Article
In: Structure, vol. 32, no. 11, pp. 1936–1951.e5, 2024, ISSN: 1878-4186.
@article{pmid39383874,
title = {Cryo-EM structures of Clostridium perfringens enterotoxin bound to its human receptor, claudin-4},
author = {Sewwandi S Rathnayake and Satchal K Erramilli and Anthony A Kossiakoff and Alex J Vecchio},
doi = {10.1016/j.str.2024.09.015},
issn = {1878-4186},
year = {2024},
date = {2024-11-01},
urldate = {2024-11-01},
journal = {Structure},
volume = {32},
number = {11},
pages = {1936--1951.e5},
abstract = {Clostridium perfringens enterotoxin (CpE) causes prevalent and deadly gastrointestinal disorders. CpE binds to receptors called claudins on the apical surfaces of small intestinal epithelium. Claudins normally regulate paracellular transport but are hijacked from doing so by CpE and are instead led to form claudin/CpE complexes. Claudin/CpE complexes are the building blocks of oligomeric β-barrel pores that penetrate the plasma membrane and induce gut cytotoxicity. Here, we present the structures of CpE in complex with its native claudin receptor in humans, claudin-4, using cryogenic electron microscopy. The structures reveal the architecture of the claudin/CpE complex, the residues used in binding, the orientation of CpE relative to the membrane, and CpE-induced changes to claudin-4. Further, structures and modeling allude to the biophysical procession from claudin/CpE complexes to cytotoxic β-barrel pores during pathogenesis. In full, this work proposes a model of claudin/CpE assembly and provides strategies to obstruct its formation to treat CpE diseases.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}